Normal medical checkups for all over 40 patients needs to include a long duration (>= 1 week) ECG screening. In the early stages of disease progression, certain types of arrhythmias and abnormal heartbeats may only surface a hand full of times a week and only under specific circumstances, making them invisible to conventional 1hr or 24hr ECG screenings.
The only way of getting this long term data, is to make the recording process itself as frictionless as possible. Enter the new generation of 3 lead halter ECG recording devices. These devices allow us to easily capture a weeks worth of ECG data, but in doing so introduce 2 new problems.
- The data contain more artifacts and have fewer channels, which means that additional training may be required to identify important arrhythmias.
- There is simply too much data.
The obvious answer is to use a ML model to identify all beats that warrant additional scrutiny and file the rest. I'm sure there are many of these models out there that do this, but as far as I know, no open-source, sate of the art, reference model for this exists, and this is a tragedy.
The developing world desperately needs to find ways to make it's existing health care resources go further. Doing ML supported ECG screening would be a great way for a country to reduce the burden on it's health care system, however health care tech startups rarely aim their products at resource constrained markets. Additionally it is usually advantageous for these solutions to be home-grown since local entrepreneurs wil often be better equipped to navigate the peculiarities of the local regulatory system. Building a business to do this is a costly and risky prospect. Would it not ne great it they did not have to start from scratch with the model as well?
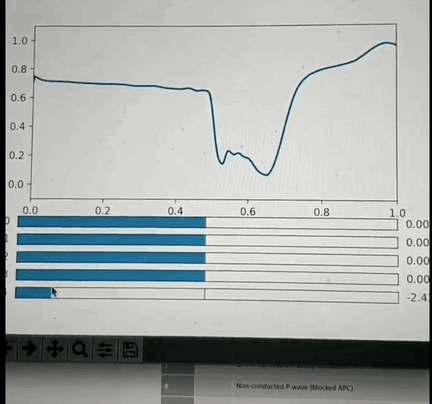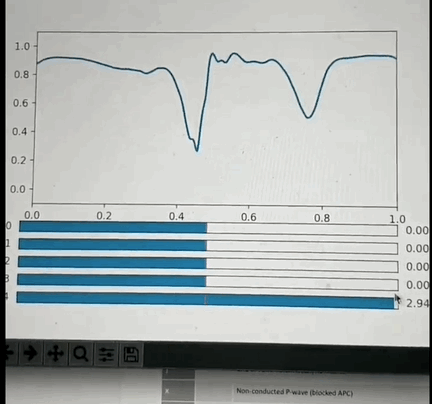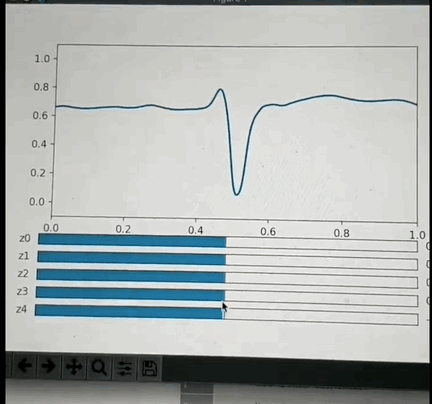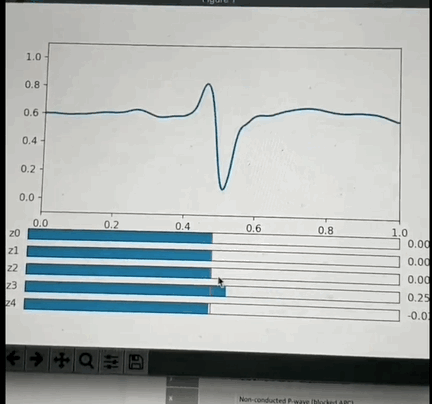
One potential approach: I've had the opportunity to look into this a little back in Oct 2019 and found that if you crop a small region (1 or 2 seconds around each beat, and center the beat), so that you have one wavelet per beat and pre-process it appropriately then you can train a VAE with a small number of latent space parameters (5 in my test case) that is able to reproduce most of the features of each beat. More interestingly: if you use some non linear clustering algorithm (I used t-SNE) to project the 5D data onto a 2-D image and color the values by the annotation you will find that each cluster of points is of the same color with less than 5% contamination. This is amazing because it means that the 5 latent space values can be combined to classify the beat pathology. What makes this blow my mind even more is the fact that the pathology labels were not used in the training of the encoder, so the model learned to cluster pathologies together based only on the similarity of the beat morphology. Above is a GIF of the decoder part of the VAE, where one can manually modify the latent space values and see the resulting waveform.